Viewing Search Results
Overview
Each of the search tools provided within the BKL generates a list of search results which are hyperlinked to the corresponding BKL reports. Individual entries can be selected for further manipulation by clicking the box at the beginning of each line, or 'Mark all on page' can be used to select all entries displayed on the current page.
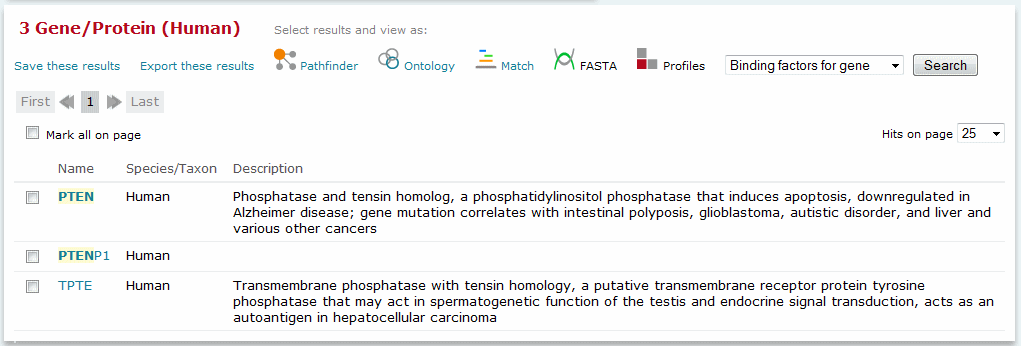
Search results
Selected entries may be further manipulated using the options described below. Please note that some options are dependent on the type of data that is present, and will not always be available.
Save these results |
Click the 'Save these results' link to save the selected (or all) entries to your personal account for future use. If you have not yet logged into the BKL you will be prompted to do so at this time. In the dialog window the default folder for the data being saved will be automatically selected and highlighted in blue. You may choose to save your data in a different folder, but please be aware that for Genes and proteins and miRNAs data sets must be stored in the default location in order to be used by the Functional Analysis and Network Analysis tools. After selecting the folder, you will assign an individual name to the stored result. If no entry has been checked and selected in the result list, the all entries from the list will be saved by default. If at least one entry has been checked and selected in the result list you will be presented with an option to save just the selected entries or to save all entries. More information about the BKL Data Management system is available here.
Export these results |
Click the 'Export these results' link to export the selected entries as a tab-delimited file. This format can be easily imported to any spreadsheet software or relational database. If no entry has been checked and selected in the result list, the result will be exported with all entries from the list.
For genes, proteins and miRNAs, additional export options are presented in a dialog window:
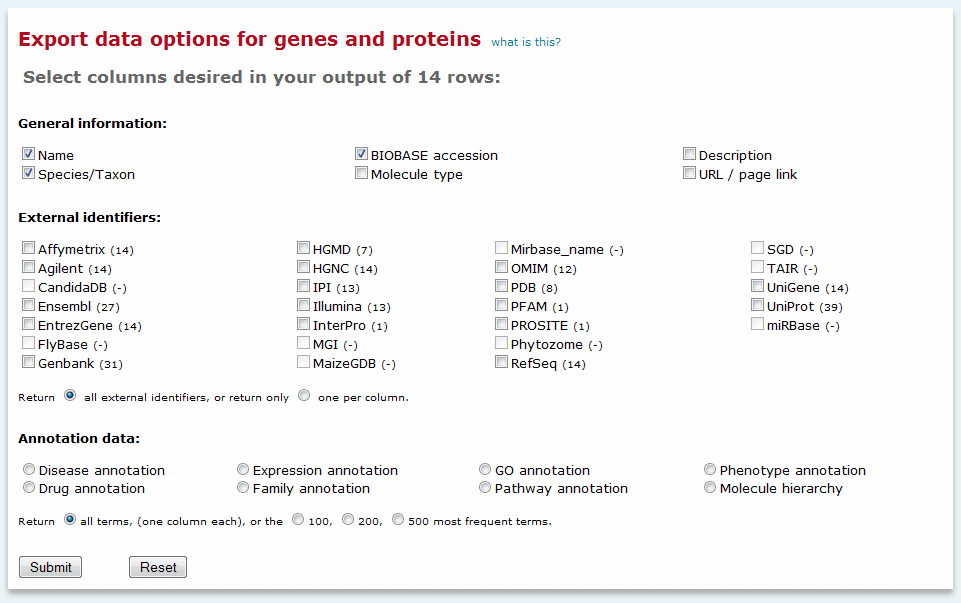
Export genes, proteins and miRNAs option
You may choose to include general information (gene or protein name, species/taxon, BIOBASE accession, a short description of the gene or protein, and a link to the relevant Locus Report), external identifiers, as well as selected controlled vocabulary terms that have been assigned to the genes or proteins. Sets of controlled vocabulary terms must be exported one at a time. For example, if you wanted to export both the disease and drug annotations for your set of results, you would first have to export one set of results and then export the other. Once exported the two sets of results can be combined into a single file as desired. In the exported results, each term is identified as a column header, with an X appearing in the row for each gene or protein that has been annotated to the term.
For external identifiers, multiple entries may be available for a given identifier. An option to return only one entry per type of external identifier selected is available by clicking the appropriate radio button.

|
Pathfinder |
Click the Pathfinder link to import the selected entries into the BKL Pathfinder visualization tool. This will open the tool and load the selected entries to the visualization canvas. If the BKL Pathfinder is already open, you will be asked if the entries should be added to the existing visualization or if the canvas should be cleared first. Please note that only genes, proteins, miRNAs, pathways and reactions are available for import into the BKL Pathfinder, and at least one entry must be selected before the button becomes active.

|
Ontology |
Click the Ontology link to import the selected entries into the Ontology Search tool where you can view all of the controlled vocabulary terms associated with your customized list. Clicking the Set Analysis tab once your genes have been imported into the Ontology Search tool will allow you to generate a report which summarizes the controlled vocabulary terms that are statistically over-represented in your list of genes.

|
Match |
Click the Match link to directly initiate a Match analysis for predicted transcription factor binding sites within the associated promoter(s) of a selected gene. Clicking the Match link opens a dialog box where you are asked to specify whether you wish to analyze all promoters or only the best-supported promoter, the sequence window to be analyzed, and the profile to be used. Results are returned in a new window. For more information about Match or about promoters, please see the Match overview and Promoter Report overview respectively. For IP-only and installed customers, please note that access to the Match feature requires login. If you have not already done so before clicking the link you will be prompted to login before proceeding to the dialog box. A subscription to the TRANSFAC module is required to access Match.

|
FASTA |
Click the FASTA link to export the sequences of the selected entries in FASTA format. Please note that only promoters, transcription factor binding sites, and ChIP fragments are available for sequence export.
For promoters, additional FASTA export options are presented. After selecting a promoter or multiple promoters from the results of an advanced promoter search, pressing "FASTA" will open a new field to the right of the search results, where you can select sequence(s) by coordinates, ranging from -10,000 base pairs to +1,000 base pairs relative to the virtual TSS. After making the appropriate selections, hit "Retrieve" to export the selected promoter sequence(s).

|
Match Profile |
Click the Match Profile link to import the selected entries into the Match Profiler tool for use in future Match analyses. Please note that only matrices are available for import into the Match Profiler tool, which requires subscription to the TRANSFAC module.
Search Within Results
The Search Within Results feature provides the ability to perform a constraint search for other data types with the selected entries. For example, it is possible to perform a search for all disease entries that are linked to the genes or proteins of a result set. You can select the data type to be queried from the drop-down menu above the "Search Within Results" button. If no entry has been checked and selected in the result list, all entries from the original result list will be used as constraint for the new query. The result of the new query will be shown in a new browser window or tab.
Results of the Topic Specific Searches (including the 'Upload a list of genes or proteins in bulk' option) and Ontology Search will present the following Search Within Result options which allow you to quickly access relevant linked data:
Gene Regulation
Binding factor for gene - treats the input as a gene or list of genes and identifies all transcription factors experimentally demonstrated to bind (high-throughput ChIP-seq data is excluded).
Gene bound by factor - treats the input as a protein or list of proteins and identifies all genes experimentally demonstrated to be bound by the protein (high-throughput ChIP-seq data is excluded).
Interacting factors - treats the input as a protein or list of proteins and identifies all transcription factors which bind to the protein or proteins.
Promoter sequence - treats the input as a gene or list of genes and retrieves the associated promoters and their associated sequences (the sequence is, by default, provide for 10,000 bp upstream of the TSS through 1,000 bp downstream of the TSS).
TF binding matrix - treats the input as a protein of list of proteins and retrieves any associated positional weight matrices.
mRNA Regulation
Regulating miRNAs - treats the input as a gene or list of genes and identifies all miRNAs experimentally demonstrated to bind the mRNAs encoded by the gene.
miRNA targets - treats the input as a miRNA or list of miRNAs and identifies all genes whose encoded mRNAs are experimentally demonstrated to be bound by the miRNA.
Functional Attributes
Disease - treats the input as a gene or list of genes and identifies all associated diseases.
Drug - treats the input as a protein or list of proteins and identifies all drugs for which the protein has been described as a target.
Interacting proteins - treats the input as a protein or list of proteins and identifies all interacting proteins.
Pathway - treats the input as a protein or protein list and identifies all pathways that the protein is a member of.
Variants - treats the input as a gene or gene list and identifies all pharmacogenomic variants that fall within the gene.
Results of the Custom Search present a different set of Search Within Result options which directly correspond to the data types described for each.