Introduction to Search Options
Overview
The Search tools presented on the start page facilitate text-based searches of the BKL to find reports that pertain to genes or proteins, miRNAs, diseases, pathways, drugs, transcription factors and positional weight matrices.
Two basic search options are provided:
The default keyword search broadly searches across all available entities (as determined by subscription) based on their ranked relevance to the search term, while topic-specific searches may be performed by clicking the 'View more search options' link. Topic specific searches typically support searching by names (common names, gene symbols), accepted identifiers or in some cases by simply browsing. Please note that disease, pathway, and drug search options are only available by subscription to the PROTEOME module, and that transcription factor and matrix search options are only available by subscription to the TRANSFAC module.
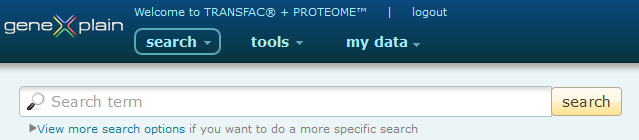
Access to topic specific searches
Results of Searches can be accessed directly, saved, exported, sent to the BKL Pathfinder (in the case of genes and proteins) or used as the input for a secondary 'Search within Results' search. The Quick Search Tools are accessible from the main search page by clicking the 'View more search options' link under the default search box.
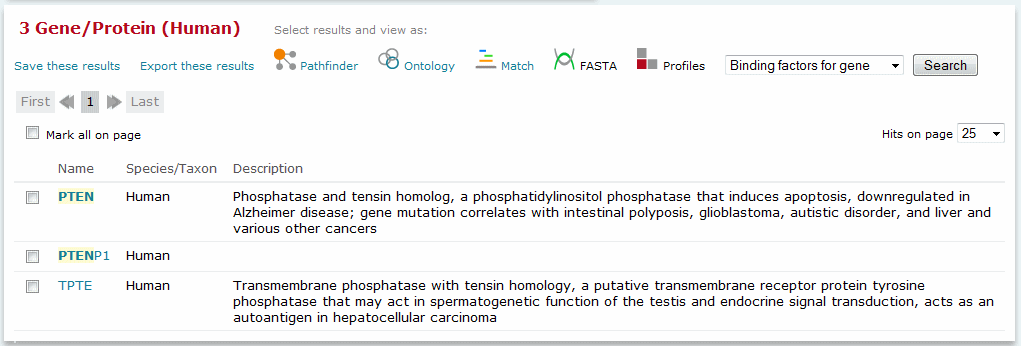
Results of a Genes or proteins topic search
Keyword Search
The keyword search allows you to search the seven main BKL topics simultaneously using a single query. The count of results found for each topic is given in the yellow summary box. Click the appropriate link to load the search results for the desired topic.
Each result returned is tagged with a phrase that describes why the result was identified by the search. A description of the tags is provided in the following table:
| Topic | Tag | Description |
|---|---|---|
| Genes and proteins | GENES_AND_PROTEINS | Represents a name or synonym of the gene/protein |
| DISEASES | Represents a name or synonym of a disease that the gene/protein is associated with | |
| DRUGS | Represents a name or synonym of a drug that targets, or is metabolized by, the gene/protein | |
| PATHWAYS | Represents a name or synonym of a pathway that the gene/protein is a member of | |
| MATRICES | Represents the name of a matrix, which represents the preferred DNA binding sequence, for the gene/protein | |
| Diseases | GENES_AND_PROTEINS | Represents a name or synonym of a gene/protein that is associated with the disease |
| DISEASES | Represents a name or synonym of the disease | |
| DRUGS | Represents a name or synonym of a drug that is under investigation for treatment of the disease in a clinical trial | |
| PATHWAYS | Represents a name or synonym of a pathway that a gene/protein associated with the disease is a member of | |
| Drugs | GENES_AND_PROTEINS | Represents a name or synonym of a gene/protein that the drug targets or is metabolized by |
| DISEASES | Represents a name or synonym of a disease that the drug is under investigation for the treatment of in a clinical trial | |
| DRUGS | Represents a name or synonym of the drug | |
| PATHWAYS | Represents a name or synonym of a pathway that a gene/protein targeted by, or metabolized by, the drug is a member of | |
| Pathways | GENES_AND_PROTEINS | Represents a name or synonym of a gene/protein that is a member of the pathway |
| DISEASES | Represents a name or synonym of a disease that is associated with a gene/protein that is a member of the pathway | |
| DRUGS | Represents a name or synonym of a drug that targets, or is metabolized by, a gene/protein that is a member of the pathway | |
| PATHWAYS | Represents a name or synonym of the pathway | |
| Transcription factors | TRANSCRIPTION_FACTORS | Represents a name or synonym of the transcription factor |
| DISEASES | Represents a name or synonym of a disease that the transcription factor is associated with | |
| DRUGS | Represents a name or synonym of a drug that targets the transcription factor | |
| PATHWAYS | Represents a name or synonym of a pathway that the transcription factor is a member of | |
| MATRICES | Represents the name of a matrix, which represents the preferred DNA binding sequence, for the transcription factor | |
| Matrices | GENES_AND_PROTEINS | Represents a name or synonym of a gene/protein (transcription factor) that the matrix represents a consensus DNA binding sequence preference for |
| DISEASES | Represents a name or synonym of a disease that is associated with a transcription factor that the matrix represents a consensus DNA binding sequence preference for | |
| DRUGS | Represents a name or synonym of a drug that targets a transcription factor that the matrix represents a consensus DNA binding sequence preference for | |
| PATHWAYS | Represents a name or synonym of a pathway that a transcription factor that the matrix represents a consensus DNA binding sequence preference for is a member of | |
| MATRICES | Represents the name of the matrix |
The ranking of individual results within a topic is determined by the number and significance of search terms matched. For example, the disease "Ovarian neoplasms" will be ranked highest in a search for "ovarian cancer" because the terms "ovarian", "cancer" and "ovarian cancer" are all names or synonyms for the disease "Ovarian neoplasms". The disease "Breast neoplasms" will be ranked lower in the same search because even though the term "cancer" is a synonym associated with the disease "Breast neoplasms", the term "ovarian cancer" is only associated with it as the synonym of a gene/protein that is associated with the disease.
If you wish to search a given topic using more specific criteria, use one of the topic searches offered under the "View more search options" link.
Topic Specific Searches
Seven Topic specific searches options are provided:
Genes and Proteins Search
To perform a search for genes and proteins, click the Genes and proteins radio button under the 'View more search options' link:
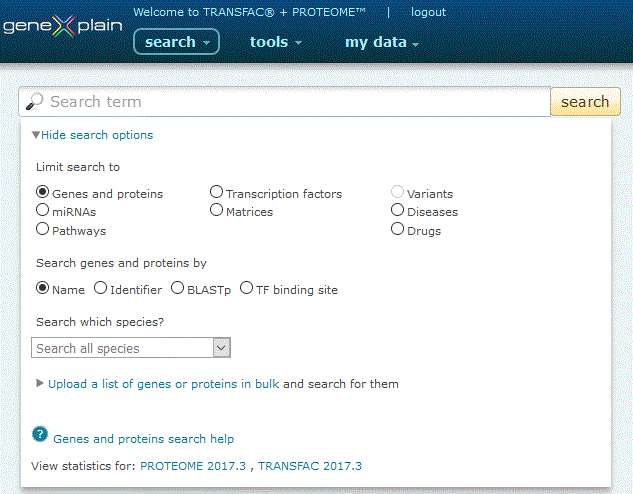
Genes and proteins search option
The Genes and proteins search provides five search options:
1. Search by name. Click the 'Name' radio button, leave the species pull-down menu set to the default to search all species or alternatively select a species to restrict your search to, type your desired search term into the search box and click the 'search' button.
The search engine queries gene and protein symbols within the name and synonyms fields (i.e. BRCA1, Ste12, NAC1). Entries are automatically wildcarded unless enclosed in double quotes (i.e. a search for BRCA will return ATBRCA1 and BRCA2 among other entries, but a search for "BRCA1" will not). Multiple names can be separated by an OR delimiter (i.e. a search for BRCA1 PSEN1 will return no results, but a search for BRCA1 OR PSEN1 will return multiple results). Once a species has been selected, a cookie will be set so that the search remembers your species preference for future searches until the Browser cache is cleared or another species has been selected.
2. Search by identifier. Click the 'Identifier' radio button, leave the identifiers pull-down menu set to the default to search all identifiers or alternatively select a type of identifier to restrict your search to (specifying the desired identifier via the pull-down menu is recommended for optimal performance), type your desired identifier into the search box and click the 'search' button.
The search engine queries BIOBASE (i.e. GN000007472, GN000003965, GN000223298) or 3rd party (i.e. Entrez Gene 672, UniProt P38398) identifiers as specified by the selection chosen from the pull-down menu. Entries are always treated as exact searches. Multiple identifiers can be separated by an OR delimiter (i.e. an EntrezGene search for 672 5663 will return no results, but an EntrezGene search for 672 OR 5663 will return BRCA1 and PSEN1), but note that all identifiers must be of the same type otherwise only the first identifier will be matched. For optimal performance it is recommended that the identifier type be specified (as opposed to searching "all" identifiers) and that no more than 50 identifiers be submitted at one time. Once an identifier has been selected, a cookie will be set so that the Quick Search remembers your identifier preference for future searches until the Browser cache is cleared or another species has been selected.
Searching for multiple names or identifiers is best done using the Upload genes and proteins in bulk option described in more detail here.
3. Search by BLASTp. Click the 'BLASTp' radio button to open a customized dialog box. Once it opens, copy and paste up to five FASTA-formatted sequences into the provided box, set the desired parameters, and click the 'blast' button.
For details of the BLAST search, click here. The BLASTp option requires a subscription to the PROTEOME module. To perform a BLAST search specific to transcription factors, click the Transcription factors radio button under the 'View more search options' link and then click the BLASTp radio button. The Transcription factors BLASTp option requires a subscription to the TRANSFAC module.
4. Search by TF binding site. Click the 'TF binding site' radio button to search for genes by a site accession (i.e. R00001) or site identifier (i.e. HS$IFI6_01).
The search engine queries the site accession and identifier fields, returning the gene which contains the site and the transcription factor which binds the site. The TF binding site option requires a subscription to the TRANSFAC module.
5. Browse by ChIP-chip and ChIP-seq experiments. Click the 'ChIP-chip and ChIP-seq experiments' link to view a list of included ChIP-chip and ChIP-seq data sets organized by transcription factor. Click the 'View Report' button for any report to read a summary of the experiment and access pre-formatted data for export and further analysis. For more information, click here.
6. Browse by factor classification. Click the 'Factor classification' link to view a list of transcription factors organized by factor classes. For more information, click here.
7. Browse by selected species. Click any of the species
links (i.e. human, mouse, Arabidopsis, etc) to export a list of
transcription factors for that species. To be included in the list,
a factor must be assigned a T accession from TRANSFAC.
The Transcription factors search option requires a subscription to
the TRANSFAC module.
8. Upload a list of genes or proteins in bulk. If you have more than a small number of genes or proteins to search for, click the 'Upload a list of genes or proteins in bulk' link to open a customized dialog box. Copy and paste your gene or proteins names or identifiers into the box provided, or specify a file to upload, following the detailed directions given here.
miRNAs Search
To perform a search for miRNAs, click the miRNAs radio button under the 'View more search options' link:
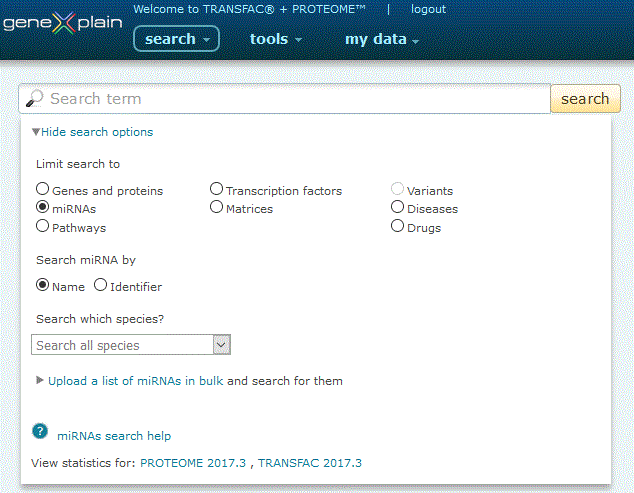
miRNAs search option
The miRNAs search provides three search options:
1. Search by name. Click the 'Name' radio button, leave the species pull-down menu set to the default to search all species or alternatively select a species to restrict your search to, type your desired search term into the search box and click the 'search' button.
The search engine queries miRNA symbols within the name and synonyms fields (i.e. MIR122, hsa-mir-155). Entries are automatically wildcarded unless enclosed in double quotes (i.e. a search for MIR122 will return MIR1224 and MIR1225 among other entries, but a search for "MIR122" will not). Multiple names can be separated by an OR delimiter (i.e. a search for MIR122 hsa-mir-155 will return no results, but a search for MIR122 OR hsa-mir-155 will return multiple results). Once a species has been selected, a cookie will be set so that the search remembers your species preference for future searches until the Browser cache is cleared or another species has been selected.
2. Search by identifier. Click the 'Identifier' radio button, leave the identifiers pull-down menu set to the default to search all identifiers or alternatively select a type of identifier to restrict your search to (specifying the desired identifier via the pull-down menu is recommended for optimal performance), type your desired identifier into the search box and click the 'search' button.
The search engine queries BIOBASE (i.e. RI000007045, RI000000949) or 3rd party (i.e. miRBase MIMAT0000646, TaqMan 002623) identifiers as specified by the selection chosen from the pull-down menu. Entries are always treated as exact searches. Multiple identifiers can be separated by an OR delimiter (i.e. a miRBase search for MIMAT0000646 MIMAT0004658 will return no results, but an EntrezGene search for MIMAT0000646 OR MIMAT0004658 will return hsa-miR-155-5p and hsa-miR-155-3p), but note that all identifiers must be of the same type otherwise only the first identifier will be matched. For optimal performance it is recommended that the identifier type be specified (as opposed to searching "all" identifiers) and that no more than 50 identifiers be submitted at one time. Once an identifier has been selected, a cookie will be set so that the Quick Search remembers your identifier preference for future searches until the Browser cache is cleared or another species has been selected.
Searching for multiple names or identifiers is best done using the Upload genes and proteins in bulk option described in more detail here.
3. Upload a list of miRNAs in bulk. If you have more than a small number of miRNAs to search for, click the 'Upload a list of miRNAs in bulk' link to open a customized dialog box. Copy and paste your miRNA names or identifiers into the box provided, or specify a file to upload, following the detailed directions given here.
Diseases Search
To perform a search for diseases, click the Diseases radio button under the 'View more search options' link:
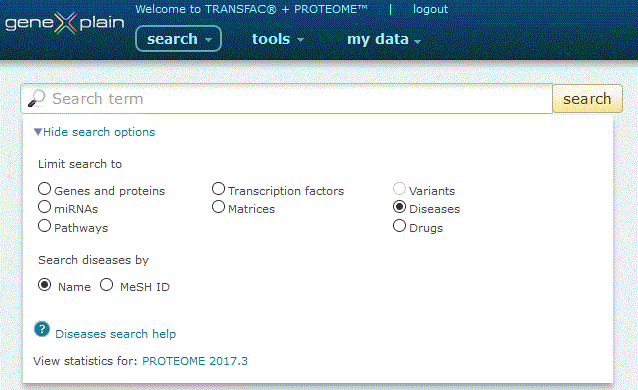
Diseases search option
The Diseases search provides two search options:
1. Search by Name. Click the 'Name' radio button, type your desired search term into the search box and click the 'search' button.
The search engine queries disease name and synonym fields (i.e. alzheimer's, lung cancer). Entries are automatically wildcarded unless enclosed in double quotes. When multiple words are entered, both must be present in the name or synonym field in order for a result to be returned (i.e. a search for lung cancer will only return records that contain both lung and cancer), unless specified by an OR delimiter (i.e. a search for lung OR cancer will return results that contain either lung or cancer).
2. Search by MeSH ID. Click the 'MeSH ID' radio button, type your desired search term into the search box and click the 'search' button.
The search engine queries the MeSH unique ID field. MeSH IDs have the format of a capital "D" plus six or nine digits, e.g. D011024 (Viral Pneunomia) or D000071243 (Zika Virus Infection).
The Diseases search option requires a subscription to the PROTEOME module.
Pathways Search
To perform a search for pathways, click the Pathways radio button under the 'View more search options' link:
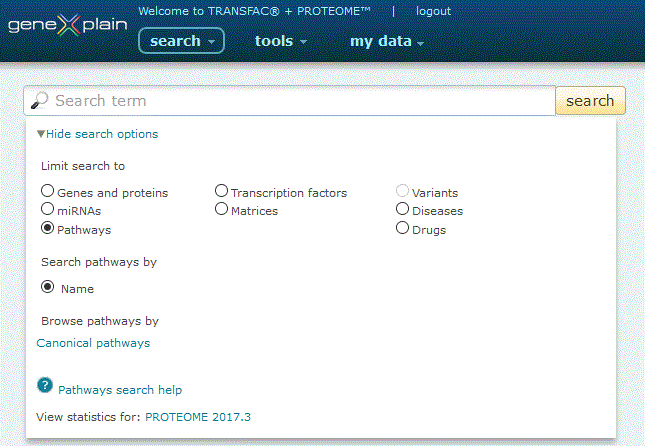
Pathways search option
The Pathways search provides two search options:
1. Search by Name. Click the 'Name' radio button, type your desired search term into the search box and click the 'search' button.
The search engine queries pathway name and synonym fields (i.e. stress-associated pathway, FAS pathway). Entries are automatically wildcarded unless enclosed in double quotes. When multiple words are entered, both must be present in the name or synonym field in order for a result to be returned (i.e. a search for stress pathway will only return records that contain both stress and pathway), unless specified by an OR delimiter (i.e. a search for stress OR pathway will return results that contain either stress or pathway).
2. Browse by Canonical pathways. Click the 'Canonical pathways' link to view a list of precompiled signaling and metabolic pathways. Click the name of the pathway to view a manually drawn graphical representation of the pathway along with a link to the complete pathway report. Alternatively, click the orange Pathfinder button to load the prepared pathway into the Pathfinder visualization tool.
The Pathways search option requires a subscription to the PROTEOME module.
Drug Search
To perform a search for drugs, click the Drugs radio button under the 'View more search options' link:
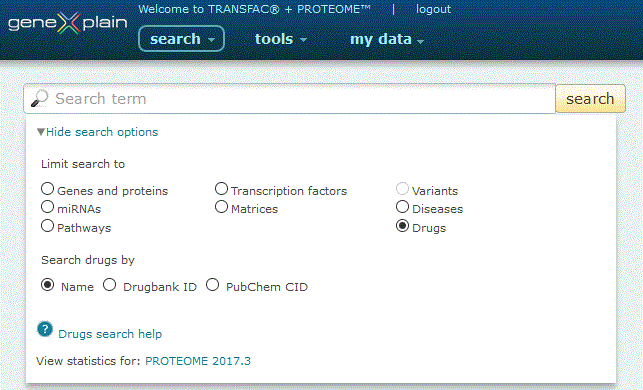
Drugs search options
The Drugs search provides three search options:
1. Search by Name. Click the 'Name' radio button, type your desired search term into the search box and click the 'search' button.
The search engine queries drug name and synonym fields (i.e. atorvastatin, benadryl). Entries are automatically wildcarded unless enclosed in double quotes. When multiple words are entered, both must be present in the name or synonym field in order for a result to be returned (i.e. a search for atorvastatin benedryl will return no results), unless specified by an OR delimiter (i.e. a search for atorvastatin OR benedryl will return multiple results).
2. Search by Drugbank ID. Click the 'Drugbank ID' radio button, type your desired search term into the search box and click the 'search' button.
3. Search by Pubchem CID. Click the 'Pubchem CID' radio button, type your desired search term into the search box and click the 'search' button.
The Drugs search option requires a subscription to the PROTEOME module.
Transcription factors Search
To perform a search for transcription factors, click the Transcription factors radio button under the 'View more search options' link:
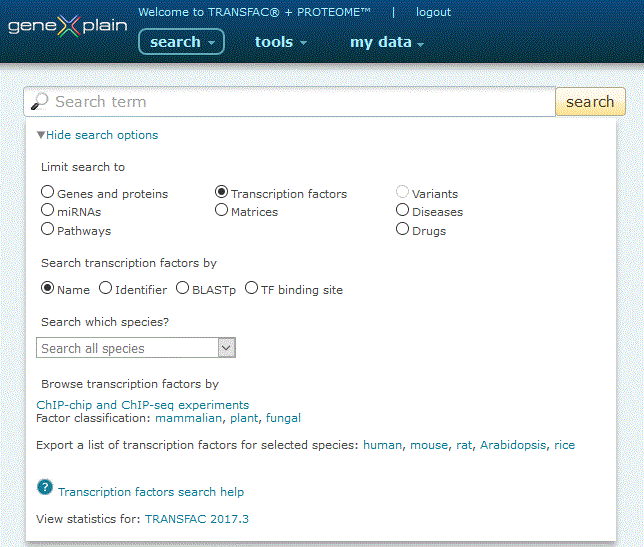
Transcription factors search option
The Transcription factors search provides seven search options:
1. Search by Name. Click the 'Name' radio button, leave the species pull-down menu set to the default to search all species or alternatively select a species to restrict your search to, type your desired search term into the search box and click the 'search' button.
The search engine queries the subset of database entries which are designated as transcription factors or other factors important for gene regulation. When the Name option is selected, the search engine queries gene and protein symbols within the name and synonyms fields (i.e. STAT1, MYC). Entries are automatically wildcarded unless enclosed in double quotes (i.e. a search for STAT will return STAT1 and STAT2 among other entries, but a search for "STAT3" will not.). Multiple names can be separated by an OR delimiter (i.e. a search for STAT1 MYC will return no results, but a search for STAT1 or MYC will return multiple results). Once a species has been selected, a cookie will be set so that the search remembers your species preference for future searches until the Browser cache is cleared or another species has been selected.
2. Search by Identifier. Click the 'Identifier' radio button, leave the identifiers pull-down menu set to the default to search all identifiers or alternatively select a type of identifier to restrict your search to (specifying the desired identifier via the pull-down menu is recommended for optimal performance), type your desired identifier into the search box and click the 'search' button.
The search engine queries BIOBASE (i.e. GN000004266, GN000000036) or 3rd party (i.e. EntrezGene 6772 , UniProt P42224) identifiers as specified by the selection chosen from the pull-down menu. Entries are always treated as exact searches. Multiple identifiers can be separated by an OR delimiter (i.e. an EntrezGene search for 6772 4609 will return no results, but an EntrezGene search for 6772 OR 4609 will return STAT1 and MYC), but note that all identifiers must be of the same type otherwise only the first identifier will be matched. For optimal performance it is recommended that the identifier type be specified (as opposed to searching "all" identifiers) and that no more than 50 identifiers be submitted at one time. Once an identifier has been selected, a cookie will be set so that the search remembers your identifier preference for future searches until the Browser cache is cleared or another species has been selected.
Searching for multiple names or identifiers is best done using the Upload genes and proteins in bulk option described in more detail here.
3. Search by BLASTp. Click the 'BLASTp' radio button to open a customized dialog box. Once it opens, copy and paste up to five FASTA-formatted sequences into the provided box, set the desired parameters, and click the 'blast' button.
The BLASTp option as part of the Transcription factors search is restricted to transcription factors. To be included in the transcription factor BLAST database, a factor must be assigned a T accession from TRANSFAC. For general details of the BLAST search, click here.
4. Search by TF binding site. Click the 'TF binding site' radio button to search for transcription factors by a site accession (i.e. R00001) or site identifier (i.e. HS$IFI6_01).
The search engine queries the site accession and identifier fields, returning the transcription factor which binds the site and the gene which contains the site. Please note that subsequent searches for 'Binding factors for gene' will take the returned gene as input, not the transcription factor.
5. Browse by ChIP-chip and ChIP-seq experiments. Click the 'ChIP-chip and ChIP-seq experiments' link to view a list of included ChIP-chip and ChIP-seq data sets organized by transcription factor. Click the 'View Report' button for any report to read a summary of the experiment and access pre-formatted data for export and further analysis. For more information, click here.
6. Browse by factor classification. Click the 'Factor classification' link to view a list of transcription factors organized by factor classes. For more information, click here.
7. Browse by selected species. Click any of the species links (i.e. human, mouse, Arabidopsis, etc) to export a list of transcription factors for that species. To be included in the list, a factor must be assigned a T accession from TRANSFAC.
The Transcription factors search option requires a subscription to the TRANSFAC module.
Matrices Search
To perform a search for matrices, click the Matrices radio button under the 'View more search options' link:
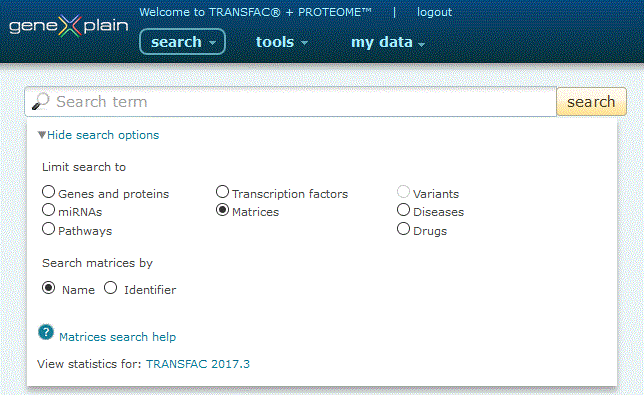
Matrices search option
The Matrices Quick Search provides two search options:
1. Search by Name. Click the 'Name' radio button, type your desired search term into the search box and click the 'search' button.
The search engine queries the library of transcription factor positional weight matrices. When the name option is selected, the accepted HGNC gene symbol is used to search for matrices for a desired transcription factor. Gene symbols (i.e. STAT1, MYC) are automatically wild carded unless enclosed in double quotes (i.e. a search for STAT will return matrices for STAT1 and STAT2 among other entries, but a search for "STAT3" will not). Multiple names can be separated by an OR delimiter (i.e. a search for STAT1 MYC will return no results, but a search for STAT1 or MYC will return multiple results).
2. Search by Identifier. Click the 'Identifier' radio button, type your desired identifier into the search box and click the 'search' button.
The search engine queries BIOBASE (i.e. M00747, M01881) identifiers for positional weight matrices.
The Matrices search option requires a subscription to the TRANSFAC module.
Search Examples and Interpretations of Different Search Types
The following examples show how searches are performed by the search engine when different search types and parameters are used.
Search Examples and Interpretation of Result
| Parameter | Search Type | |
|---|---|---|
| Search Term | Interpretation of the result | |
| Name Search | ||
| Gene / Protein and Transcription Factor | p53 single search term |
wildcarded as %p53%, gene and protein names and synonyms |
|
"p53" single search term in double quotes |
exact term is searched, gene and protein names and synonyms | |
| p53
brca1 2 search terms separated by space |
wildcarded as %p53% and %brca1%, gene and protein names and synonyms containing both search terms | |
| "p53"
"brca1" 2 search terms both in double quotes |
exact terms are searched, gene and protein names and synonyms containing one of both search terms | |
| "p53
brca1" 2 search terms together in double quotes |
exact term is searched, gene and protein names and synonyms containing both search terms together | |
| p53 OR
brca1 2 search terms separated by an OR operator |
wildcarded as %p53% OR %brca1%, gene and protein names and synonyms containing one of both search terms | |
| "p53"
OR "brca1" 2 search terms both in double quotes and separated by an OR operator |
exact terms are searched, gene and protein names and synonyms containing one of both search terms | |
| p53 AND
brca1 2 search terms separated by an AND operator |
wildcarded as %p53% AND %brca1%, gene and protein names and synonyms containing both search terms | |
| "p53"
AND "brca1" 2 search terms together in double quotes |
2 search terms both in double quotes and separated by an AND operator | |
| Disease |
Asthma single search term |
wildcarded as %Asthma%, disease names and synonyms |
|
"Asthma" single search term in double quotes |
exact term is searched, disease names and synonyms | |
| Common
Cold 2 search terms separated by space |
wildcarded as %Common% and %Cold%, disease names and synonyms containing both search terms | |
| "Lung"
"Neoplasms" 2 search terms both in double quotes |
exact terms are searched, disease names and synonyms containing one of both search terms | |
| "Common
cold" 2 search terms together in double quotes |
exact term is searched, disease names and synonyms containing both search terms together | |
| Asthma
OR Psoriasis 2 search terms separated by an OR operator |
wildcarded as %Asthma% OR %Psoriasis%, disease names and synonyms containing one of both search terms | |
|
"Asthma" OR "Psoriasis" 2 search terms both in double quotes and separated by an OR operator |
exact terms are searched, disease names and synonyms containing one of both search terms | |
| Asthma
AND Psoriasis 2 search terms separated by an AND operator |
wildcarded as %Asthma% AND %Psoriasis%, disease names and synonyms containing both search terms | |
|
"Asthma" AND "Psoriasis" 2 search terms both in double quotes and separated by an AND operator |
exact terms are searched, disease names and synonyms containing both search terms | |
| Pathway |
Caspase single search term |
wildcarded as %Caspase%, pathway name and synonym |
|
"Glycolysis" single search term in double quotes |
exact term is searched, pathway name and synonym | |
| Caspase
network 2 search terms separated by space |
wildcarded as %Caspase% and %network%, pathway name and synonyms containing both search terms | |
|
"glycolysis" "AR pathway" 2 search terms both in double quotes |
exact terms are searched, pathway name and synonym containing one of both search terms | |
| "NOS
regulation" 2 search terms together in double quotes |
exact term is searched, pathway name and synonym containing both search terms together | |
| Caspase
network OR AR Pathway 2 search terms separated by an OR operator |
wildcarded as %Caspase% AND %network% OR %AR% AND %Pathway%, pathway name and synonym containing one of both search terms | |
|
"Caspase network" OR "AR Pathway" 2 search terms both in double quotes and separated by an OR operator |
exact terms are searched, pathway name and synonym containing one of both search terms | |
| Caspase
network AND AR Pathway 2 search terms separated by an AND operator |
wildcarded as %Caspase% AND %network% AND %AR% AND %Pathway%, pathway name and synonym containing both search terms | |
|
"Caspase network" AND "AR Pathway" 2 search terms both in double quotes and separated by an AND operator |
exact terms are searched, pathway name and synonym containing both search terms | |
| Drug |
Cetrizine single search term |
wildcarded as %Cetrizine%, drug name, synonyms and brand name |
|
"Cetrizine" single search term in double quotes |
exact term is searched, drug name, synonyms and brand name | |
|
Ethacrynic acid 2 search terms separated by space |
wildcarded as %Ethacrynic% and %acid%, drug name, synonyms and brand name containing both search terms | |
|
"Ibuprofen" "Aspirin" 2 search terms both in double quotes |
exact terms are searched, drug name, synonyms and brand name containing one of both search terms | |
|
"Ethacrynic acid" 2 search terms together in double quotes |
exact term is searched, drug name, synonyms and brand name containing both search terms together | |
|
Ibuprofen OR Aspirin 2 search terms separated by an OR operator |
wildcarded as %Ibuprofen% OR %Aspirin%, drug name, synonyms and brand name containing one of both search terms | |
|
"Ibuprofen" OR "Aspirin" 2 search terms both in double quotes and separated by an OR operator |
exact terms are searched, drug name, synonyms and brand name containing one of both search terms | |
|
Ibuprofen AND Aspirin 2 search terms separated by an AND operato |
wildcarded as %Ibuprofen% AND %Aspirin%, drug name, synonyms and brand name containing both search terms | |
|
"Ibuprofen" AND "Aspirin" 2 search terms both in double quotes and separated by an AND operator |
2 search terms both in double quotes and separated by an AND operator | |
| Identifier Search | ||
| Gene / Protein and Transcription Factor |
GN000007472 single Identifier |
exact Identifier is searched, according to the pull-down menu |
|
"GN000007472" single Identifier in double quotes |
exact Identifier is searched, according to the pull-down menu | |
|
GN000007472 GN000007445 2 Identifier separated by space |
exact Identifier is searched, according to the pull-down menu, returns all entries matching to one of both identifiers | |
|
"GN000007472" "GN000007445" 2 Identifier both in double quotes |
exact Identifier is searched, according to the pull-down menu, returns all entries matching to one of both identifiers | |
|
"GN000007472 GN000007445" 2 Identifier together in double quotes |
exact Identifier is searched, according to the pull-down menu, returns all entries matching to one of both identifiers | |
|
GN000007472 OR GN00007445 2 Identifier separated by an OR operator |
exact Identifier is searched, according to the pull-down menu, returns all entries matching to one of both identifiers | |
| %7472%
OR %1445% 2 Identifier both wildcarded and separated by an OR operato |
according to the pull-down menu, returns all entries matching to one of both wildcarded identifiers | |
|
"GN000007472" OR "GN000007445" 2 Identifier both in double quotes and separated by an OR operator |
exact Identifier is searched, according to the pull-down menu, returns all entries matching to one of both identifiers | |
Viewing Results
Results of Gene/Protein and Transcription Factor Name searches
are a list of genes/proteins. The hits in the results list are
sorted first by exact match then alphanumerically by species with
preference given to human, mouse, rat, and S. cerevisiae. Click the
linked name to access the relevant Locus Report.
Results of Gene/Protein and Transcription Factor Identifier
searches are a list of genes/proteins (depending on the type of
identifier used as input and availability of data, the result can
contain either genes or proteins). The hits in the results list are
exact hits and are sorted alphanumerically. Click the linked name
to access the relevant Locus Report.
Results of miRNA Name searches are a list of mature miRNAs. The
hits in the results list are sorted first by exact match then
alphanumerically by species with preference given to human, mouse,
and rat. Click the linked name to access the relevant miRNA
Report.
Results of miRNA Identifier searches are a list of mature
miRNAs. The hits in the results list are exact hits and are sorted
alphanumerically. Click the linked name to access the relevant
miRNA Report.
Results of Disease searches are a list of diseases. The hits in the results list are sorted alphabetically. Click the linked name to access the relevant Disease Report.
Results of Pathway searches are a list of pathways. The hits in the results list are sorted alphabetically. Click the linked name to access the relevant Pathway Report.
Results of Drug Quick Searches are a list of drugs. The hits in the results list are sorted alphabetically. Click the linked name to access the relevant Drug Report.
Results of Matrix Quick Searches are a list of matrices and associated consensus sequences. Click the linked accession to access the relevant Matrix Report,
Results of Variants Quick Searches are a list of variants and associated genes, diseases, and drugs and observed phenotypes. Click the linked dbSNP ID or BIOBASE accession to access the relevant Variant Report. To jump to the section of the Variant Report that is specific to a phenotype of interest, click the 'view details' link next to the desired phenotype.
Click here to learn about the options available for further manipulation of Search Results.