Set Analysis
Overview
The Set Analysis feature within the Ontology Search tool identifies curated vocabulary terms that are unusually common within a pool of selected genes, relative to the overall occurrence of those terms in the complete subscribed set of genes. For instance, if a user performed an experiment to detect genes expressed in liver cells, and entered that set of genes via the main search's "Upload a list of genes or proteins in bulk" option, those vocabulary terms related to liver cell gene expression would be expected to be over-represented. While the term "EX:liver" might have been curated to 20% of the total genes with Expression curation in the user's subscribed volume of the BKL, the number of genes annotated to "EX:liver" among the user's input set of genes might be closer to 95% or higher, making that term dramatically over-represented. Likewise, if a user focuses on the term "MF:motor activity", the resulting set of genes will also show a dramatic over-representation of curated terms such as "CC:cytoskeleton", "DO:Kinesin motor domain", and "MF:ATPase activity" due to their common assignment to many of these same genes.
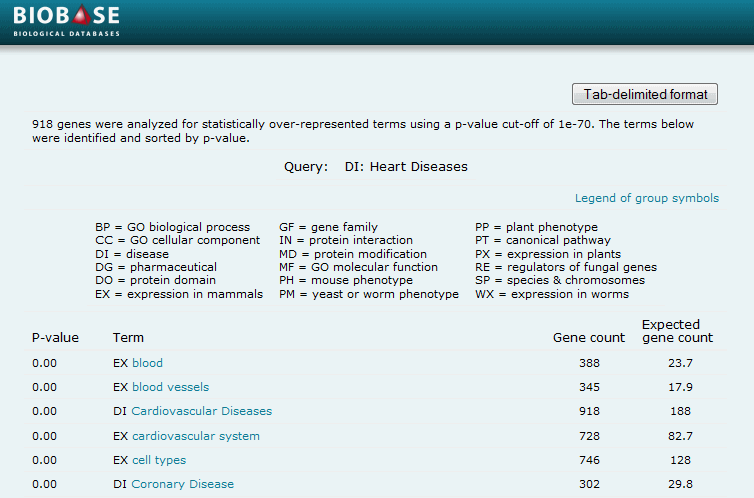
When the Set Analysis feature within the Ontology Search tool is clicked, P-values are displayed in the driller. To return a list of terms that are significantly over-represented in the set of selected genes, specify the desired P-value from the pull-down menu and specify whether the terms should be grouped by proximity (terms with a parent-child relationship are grouped together) or ranked by P-value. After clicking Show report, the terms will be presented in a pop-up window along with the P-value, determined gene count, and expected gene count. All terms are hyperlinked such that clicking the term takes you directly to that term in the Driller.
Instructions for using the Set Analysis feature of the Ontology Search tool to calculate statistics for controlled vocabulary terms are provided below. Click here for detailed information about how the term statistics are calculated.
Please note: The Set Analysis option will only become available as a link under the driller window once a set of genes has been imported into the Ontology search tool from the results of another search or as soon as the first search event has been executed..
Please note: For simplicity and brevity, the documentation uses genes as the focus of the Set Analysis feature, but diseases and drugs can also be made to be the focus of by selecting either from the pull-down menu at the top right corner of the tool..
Using the Set Analysis Feature of the Ontology Search tool to Calculate Controlled Vocabulary Term Statistics
To perform a Set Analysis for a set of selected genes:
-
Select a set of genes of interest, either by entering a list of genes via the "Upload a list of genes or proteins in bulk" option of the main search or searching for a specific term or set of term within the Ontology Search tool. If you enter a list of genes via the "Upload a list of genes or proteins in bulk" option of the main search, you must click the Ontology link to import the list of matched genes into the Ontology Search tool.
-
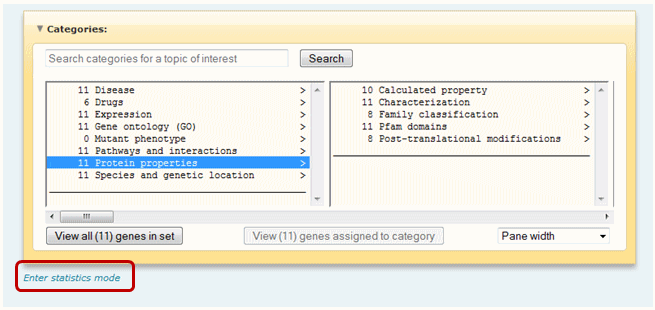
Click on the "Enter statistics mode" link below the driller window. The driller will refresh with P-values next to each term.
-
Once the "Enter statistics mode" link is clicked, a set of options will be provided. Specify the desired P-value to apply from the pull-down menu and whether you wish to see related terms grouped together (in this case select the "Proximity and p-value (group by related terms)" radio button) or listed from most significant to least significant (in this case select the "P-value" radio button).
-

-
Click the "Show report" button. From the generated report, click the Tab-delimited format button to view the results in tab-delimited form. To obtain a list of genes annotated to a term of interest, click the term link in the report which take you directly to the term in the window. Click the "View genes assigned to category" button to return the list of annotated genes and then click the "Export these results" link to export the list of annotated genes along with their term assignments.