Upload Genes and Proteins in Bulk
Overview
To upload a list of genes or proteins by name or identifier in bulk, thereby making them available for further analysis, use the "Upload a list of genes or proteins in bulk" option in the "Genes and proteins" topic search.
The section below contains:
- Instructions for uploading data
- Accepted identifiers for uploading data
- Guidelines for uploading data
Uploading data
The "Upload a list of genes or proteins in bulk" option is accessed by clicking the "View more search options" link on the main BKL page, selecting the "Genes and proteins" radio button, then clicking the "Upload a list of genes and proteins in bulk" link.
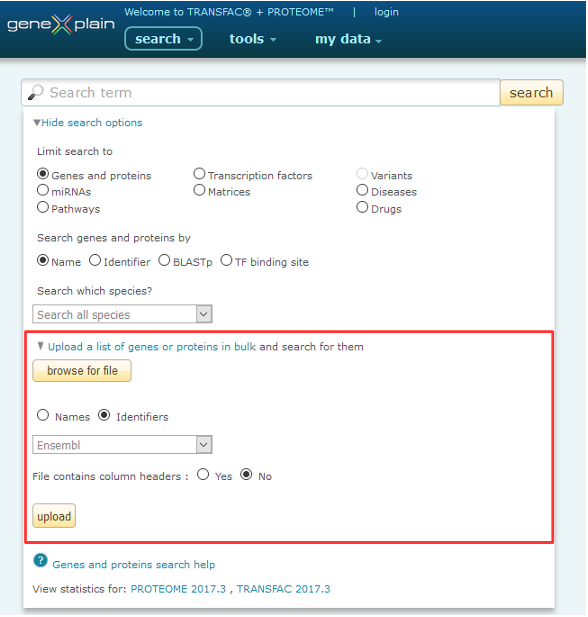
Uploading a list of genes or proteins into the BKL
To upload your own list of genes or proteins:
1. Click the "Upload a list of genes or proteins in bulk" link to make the data entry form appear.
2. Enter your data by one of the following methods:
- Copying and pasting a list of genes from a file in the text
box.
- Browsing for a tab-delimited text file.
- Manually entering IDs or gene symbols, one per line in the text box.
See the guidelines below for tips on entering data.
3. Specify whether you are entering Names (symbols) or Identifiers by selecting the appropriate radio button. Accepted data types are listed in the table below.
- When uploading Names (symbols), you will be prompted to specify whether you are uploading a list of genes and proteins or miRNAs, the species, and whether you want to prefer matches to gene names over synonyms.
- When uploading Identifiers, you will be prompted to specify the identifier type. accepted IDs are listed in the table below.
4. Click the "upload" button. The matched genes will appear in the Search Results list.
Although only one sequence is displayed on the Locus Report for each gene, multiple sequences may be internally associated with the encoded protein(s) and may contribute to the matching of accessions/IDs.
Unmatched input data will be noted in red near the bottom of the search results output. Genes that match multiple input data will be listed only once. The search results can now be manipulated in multiple ways for further analysis including:
-
Clicking the "Save these results" link allows you to store your list for use in the future.
-
Clicking the "Export these results" link allows you to export your list supplemented with controlled vocabulary terms of your choice (for example, you may choose to export the curated disease assignments for all of the genes in your list).
-
Clicking the Pathfinder link allows you to import the selected entries into the BKL Pathfinder tool in order to build out networks involving your genes or proteins (please note that at least one entry must be selected in order for the Pathfinder button to become activated).
-
Clicking the Ontology link allows you to import your list into the Ontology Search tool where you can view all of the controlled vocabulary terms associated with your customized list. Clicking the "Enter statistics mode" link once your genes have been imported into the Ontology Search tool will allow you to generate a report which summarizes the controlled vocabulary terms that are statistically over-represented in your list of genes.
Please note: Click here for details about known browser limitations for data input.
Accepted identifiers (IDs)
The following IDs are accepted by the BKL:
| ID Type | Species Included | Examples of Valid Format |
|---|---|---|
| Affymetrix probe IDs | Human, mouse, rat | 208445_s_at returns human BAZ1B |
| Agilent probe IDs | Human, mouse, rat | A_44_P118929 returns rat p53 |
| CandidaDB IDs | C. albicans | CA4342 returns Candida albicans ABD1. |
| Ensembl IDs | Human, mouse, rat | ENSG00000091831 returns human ESR1. |
| Entrez Gene IDs | Human, mouse, rat, Arabidopsis, maize, rice | 1132 returns human CHRM4. |
| GenBank Accession | All | Includes nucleotide IDs, protein IDs, and GI numbers. For example, NM_000741.1, NP_000732.1, GI:4502821, and GI:4502820 all return human CHRM4. |
| HGNC IDs | Human | 1585 returns human cyclin D3 |
| IGRSP IDs | IGRSP IDs | Os03g0707600 returns rice SLR1 |
| IPI IDs | Human, mouse, rat | IPI00009225 returns human syntaxin-8 |
| MGI IDs | Mouse | 88350 returns mouse Cdc25C |
| OMIM IDs | Human, mouse | 601128 returns human H3F3A and H3F3B |
| Phytozome | Soybean, sorghum | Glyma08g22190 picks up itself |
| PubMedIDs | All | 7642747 returns human CHRM1, CHRM2, CHRM3, CHRM4, CHRM5. |
| SGD IDs | S. cerevisiae | S000006358 returns
S. cerevisiae PIN3; S000005160 returns S. cerevisiae RAP1 |
| TAIR IDs (AGI Identifiers) |
Arabidopsis | AT1G69120 returns Arabidopsis AP1 |
| TIGR Locus IDs | rice | LOC_Os03g17700 returns rice MPK5 |
| TRANSFAC Factor IDs | All | T00261 returns human ESR1. |
| TRANSFAC System Gene IDs | All | G003925 returns human ESR1. |
| TRANSPATH Molecule IDs | All | MO000019431 returns human ESR1. |
| Unigene IDs | Human, mouse, rat | 248100 returns human CHRM4. |
| UniProt IDs | All | P08173 returns human CHRM4. |
| WormPep IDs | C. elegans | CEO1784 returns C. elegans sem-5. |
Precomputed matches of public accessions/identifiers (IDs) to genes within the BKL are derived as follows:
- Matches to GenBank nucleotide and protein IDs for human, mouse,
and rat proteins are determined by performing BLAST searches with
each sequence against dbEST (human), gb(PRI) Primate, RefSeq, gbHTC
(high-throughput cDNA collection), and dbROD (rodent
collection).
- Matches to GenBank nucleotide and protein IDs for S. cerevisiae, S. pombe, C. elegans, and the 18 species
contained in MycoPathPD are determined by the sequences assigned to
each protein in the database.
- Matches to UniProt IDs for all species are determined by the
sequences assigned to each protein in the database.
- Matches to Entrez Gene IDs and UniGene IDs for human, mouse,
and rat are determined by the sequences assigned to each protein in
the database.
- Matches to SGD (for S. cerevisiae proteins), CandidaDB (for
C. albicans proteins), and
WormPep (for C. elegans
proteins) IDs are determined by sequences assigned to each protein
in the database.
- Matches to AGI identifier (Arabidopsis) or IGRSP (rice) are
determined by sequences assigned to each protein in the
database.
Note: AGI and IGRSP IDs should be uploaded using "Gene or Protein Names".
- Matches to TIGR ID (rice) are derived from mapping to IGRSP
models by RAP-DB.
Note: these IDs should be uploaded using "Gene or Protein Names".
- Matches to Phytozome identifiers (soybean and sorghum) are
determined by sequences assigned to each protein in the
database.
Note: these IDs should be uploaded using "Gene or Protein Names".
- Matches to other identifier types are either determined from
sequences assigned to each protein in the database or through
manual curation.
In addition, select proteins can be searched with identifiers from the following databases: PDB, PFAM, PROSITE, Interpro, IPI, PIR, and MaizeGDB.
Guidelines for uploading data
Keep the following guidelines in mind when uploading your data:
- The input form accepts any synonym for a gene or protein that
has been curated from the published literature, but not all
synonyms may be represented within the BKL.
- The form is case-insensitive for gene symbol searches.
- Because more than one protein may share a gene symbol that you
entered (i.e., matches one or more synonyms in the chosen species),
you must specify whether you wish to "Match all gene symbols",
"Match primary symbols only", or "Match synonyms only if
necessary". If you choose to "Match all gene symbols", all
synonym-matched genes are returned in addition to the primary
gene.
- When entering gene symbols, you must designate a species by
doing either of the following:
- Select a species from the pull-down menu.
- Enter the species name after each gene symbol, separating the
gene symbol and species name by either a tab or "@@".
- Select a species from the pull-down menu.
- If entering your data using a tab-delimited text file, make sure the gene symbol is in the first tab position and the species name (if more than one species is present) is in the second position. If using accession numbers of any type, make sure they are in the first tab position.