Building Queries with the Ontology Search Tool
Overview
Genes, miRNAs, diseases, and drugs in the BKL are characterized using controlled vocabulary (property terms). For genes, these terms describe, among other topics, molecular function, biological role, localization, expression pattern, mutant phenotype, disease relevance, chromosomal location, and the presence of conserved domain motifs. Genes are further characterized by a set of calculated properties (presented as ranges), which include protein length, weight, and isoelectric point. In its gene-centric mode, the Property Search allows you to find all genes associated with selected controlled vocabulary terms or combinations of terms. The Ontology Search also allows you to view all the controlled vocabulary terms associated with a set of genes that you enter. In its disease-centric mode, the Ontology Search allows you to find all diseases associated with selected controlled vocabulary terms or combinations of terms, while in its drug-centric mode, the Ontology Search allows you to find all drugs associated with selected controlled vocabulary terms or combination of terms.
Detailed descriptions of the property categories used in the Ontology Search Tool are provided here.
Building a query
Building a query with the Ontology Search tool involves the following steps:
1. Select the entity to be searched
3. Add optional OR or qualifier terms if desired
For detailed step-by-step examples of queries, click the "show this example" options provided in the introductory text above the entity pull-down menu.
1. Select the entity to be searched
The Ontology Search tool provides the option to search for genes, miRNAs, diseases or drugs. Specify the type of entity using the "Search for" pull-down menu. When an entity is selected its respective hierarchy of search terms will be automatically loaded in the driller window.
Please note: Loading the vocabularies can take up to a few minutes but typically takes less than 30s, depending on the browser and connection speed.
For simplicity and brevity, the remainder of the documentation will use genes as the focus of the search and will only refer to diseases and drugs as the focus of the search when a unique function is involved.
2. Select the search term
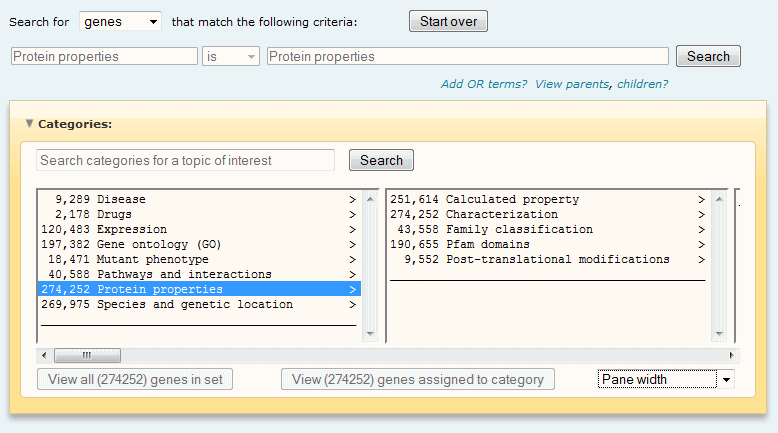
Ontology Search term selection
Search terms are selected from the driller window by browsing or by using category search. Key features and functions of the driller window include:
- The driller provides an overview of the hierarchy of terms,
enabling you to browse through the driller hierarchies to find a
search term of interest.
- The Driller root categories (all the controlled vocabularies
and other gene qualifiers - Characterization, Species, etc) are
displayed in the first panel. Selecting a term will display the
next level of terms in the panels toward the right. As you move
toward the right, the terms become more specific. Click here for more information about
this feature.
- Numbers next to each term show how many distinct genes in the
current found set are assigned to a term or its children.
- Most terms in the driller are accompanied by the greater than
symbol (>), indicating that the term has
children. If that symbol is missing, the term has no children (in other
words, it is a terminal node).
- The currently selected term is highlighted and appears
in the active query line above the driller window.
- When you click the "Search" button in the active query line to
execute the search, the Ontology Search Tool uses the currently
selected term in the Driller to search the installed BKL or custom
gene set for all associated genes. The score, or number, of these
genes associated with each vocabulary term is then returned to the
driller.
- Initially, the counts or scores in the driller reflect all
available genes, but after performing a search, the counts or
scores reflect only those genes associated with the searched term.
Sequential search events further narrows the set of genes and the
number of associated terms.
- You can choose to have the terms in the driller sorted in ascending alphabetical order or in descending order by score (number of genes associated with a controlled vocabulary term or any child terms of the selected controlled vocabulary term). By default, the driller sorts terms alphabetically. To change the sorting criteria, select the desired option from the "Pane width" pull-down menu at the bottom right of the driller window.
- You can customize the width of the panes in the driller window by selecting the desired incremental change in the "Pane width" pull-down menu at the bottom right of the driller window.
Category search
Although browsing the hierarchies of terms is useful for becoming familiar with their contents, the category search option is best used when you want to find a specific term. Type the desired term into the text box within the driller window and click the "Search" button. The results will appear in a pull-down menu next to the search button, from which you can select the term you want to use for the search. Alternatively, select the "Pop-up results" option in the pull-down menu to view the search results in a separate window. When a term is selected, the driller will automatically highlight the term in blue and it will become displayed in the active line of the query.
Keep the following guidelines in mind when performing searches for property terms.
- All words have an implicit wild card on each side so partial
terms can be used. Click here for a
description of accepted wild cards and further
information.
- Spaces count in this search, enabling you to search for complex
terms.
- Search for the most specific applicable term. For example, if you are interested in calmodulin-dependent protein kinases, a search for kinase yields 258 different Gene Ontology (GO) terms that contain the word kinase, while a search for calmodulin yields 9 different Gene Ontology (GO) terms that contain the word calmodulin.
Is / Is not
Use the is/is not pull-down menu which precedes the term in the active line of the query to specify whether you want to include all genes meeting the search criteria in your search results (if so, specify "is") or whether you want to exclude all genes meeting the search criteria from your search results (if so, specify "is not"). Note that the "is not" option only becomes available after at least one "is" search has been performed.
The controlled vocabulary hierarchy for each term in the list is designated by the codes listed in the table below. Additional information about the Property Categories is provided here.
Codes for Gene Property Categories in the Ontology Search Tool
| Code | Property Category |
|---|---|
| MF | GO Molecular Function |
| BP | GO Biological Process |
| CC | GO Cellular Component |
| GF | Family classification |
| CH | Characterization |
| DO | Protein Domain |
| EX | Expression |
| DI | Disease |
| DG | Pharmaceuticals |
| IN | Interaction with other proteins |
| PH | Phenotype |
| EV | Evidence |
| OL | Orthologs related to current set |
| SP | Species, includes chromosomal location |
| RG | Protein length, weight, and isoelectric point ranges |
| MD | Protein modifications (fungi, worm) |
| RE | Regulators of fungal genes |
| TF | Regulatory factors and targets |
| PT | Pathway |
Codes for Disease Property Categories in the Ontology Search Tool
| Code | Property Category |
|---|---|
| GA | Gene associations |
| DI | Co-occurring conditions and diseases |
| CL | Clinical trial status |
| MO | Has mouse model |
| GO | Affected biological process |
| EX | Affected cell types, tissues |
Codes for Drug Property Categories in the Ontology Search Tool
| Code | Property Category |
|---|---|
| DC | Drug category (DrugBank) |
| DT | Drug toxicity bioassays |
| PT | Protein targets |
| CT | Clinical trial status |
| DI | Disease connections |
| PW | Pathways |
| EM | Enzyme metabolizers |
Codes for miRNA Property Categories in the Ontology Search Tool
| Code | Property Category |
|---|---|
| DI | Disease |
| EX | Expression |
| MF | GO Molecular Function |
| BP | GO Biological Process |
| CC | GO Cellular Component |
| FM | miRNA properties |
3. Add optional OR or qualifier terms if desired
Add OR terms?
Once a term has been selected, you may optionally click the "Add OR terms?" link which always appears below the active query line. Doing so will add a new, linked active query line which can be used to specify a second term to be combined with the first. For example, you may want to search for all genes that are expressed in either the prostate OR ovary. Additional terms may be specified by clicking the plus button that appears next to the active query line. Terms may be removed by clicking the minus button that appears next to a query line.
Add optional qualifiers?
Certain hierarchies support the optional addition of qualifier terms which may be applied in order to further increase the specificity of the search term. For example, you may be interested in identifying all genes that are expressed in the liver but want to specifically limit your results to those genes that have been shown to be expressed in the liver using Northern analysis as the technique. Or you may be interested in identifying all genes that are associated with Alzheimer's disease but want to specifically limit your results to those genes that have been shown to be associated with Alzheimer's disease due to an increase in enzymatic activity. Once a term has been selected in a hierarchy that supports this feature, the "Add optional qualifiers?" link will appear below the active query line. Clicking the link will add a new, linked active query line which can be used to specify the desired qualifying term from the set of terms displayed in red text (if you don't see terms in red text within the driller window, scroll back to the root pane). Additional terms may be specified by clicking the plus button that appears next to the active query line. Terms may be removed by clicking the minus button that appears next to a query line.
View parents, children?
The "View parents, children?" link which always appears below the active query line does not influence the query, but provides a convenient mechanism for viewing the terms as a vertical tree in the pop-up that opens in a new window. Clicking a term within the pop-up automatically highlights the term in the driller window.
4. Execute the search
Once you have finished selecting your search term and optional OR or qualifier terms, click the "Search" button to execute the search. The Ontology Search tool searches the installed BKL (or custom gene set if you have imported a list of genes into the Ontology search tool from a set of search results) for all associated genes. The "Search" button changes to "Remove" and the count of genes returned by the search is displayed next to the button. At the same time, the driller window refreshes to display the count of genes within the result set that are associated with each of the vocabulary terms in the driller window. A new active query line is added which can be used to select the next search term and narrow the set of genes further by repeating steps 2 and 3 if desired.
At any point during query building, any "Remove" button can be clicked to remove the search term from the query. In such cases the count of genes in the result set will be recalculated and the drill window will be refreshed to reflect the roll back. To begin a query from scratch, click the "Start over" button.
5. View search results
At any time while building your query you may choose to view the list of genes meeting the specified criteria by clicking the "View genes assigned to category" and "View all genes in set" buttons. These buttons will initially appear as unclickable and will remain in that state until the number of genes is reduced to less than 20,000. Once that threshold has been reached, the "View genes assigned to category" button may be used at any point while browsing through the terms in the driller window. The "View all genes in set" is more restricted and may only be used after a search has been executed, always returning the set of genes specified by the query. Each result in the list will be hyperlinked to its respective Locus Report.
Once a list of genes has been returned, additional options are made available including the ability to save and/or export the list of genes/diseases/drugs, to load selected genes into the Pathfinder visualization tool as well as to perform selected secondary searches using the results of your query as input. Learn more about options available for search results here.