Property Details
Overview
This section describes the root categories in the driller that allow you to focus on several different types of properties and retrieve associated BKL reports. Categories are listed below, with hyperlinks to their corresponding descriptions.
Gene Controlled Vocabulary Property Categories (and their respective codes)
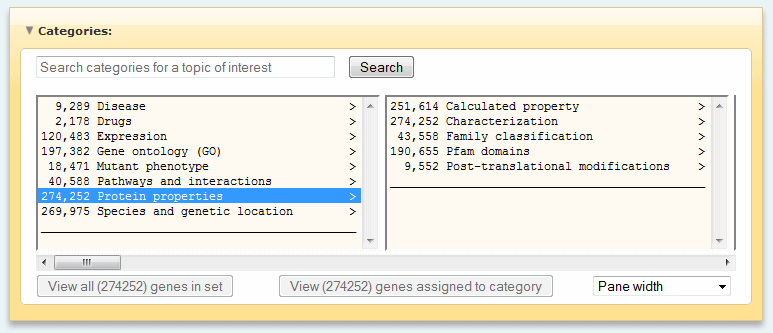
Gene properties
- Gene ontology (GO) (MF, BP, CC)
- Expression (EX)
- Disease (DI)
- Drugs (DG)
- Mutant phenotype (PH)
- Pathways and interactions
- Interaction with other proteins
(IN)
- Pathways (PT)
- Interaction with other proteins
(IN)
- Species and genetic location
(SP)
- Protein properties
Disease Controlled Vocabulary Property Categories (and their respective codes)
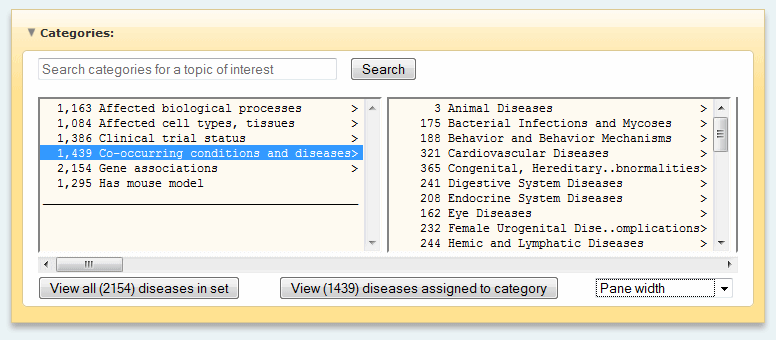
Disease properties
- Gene associations
(GA)
- Co-occuring conditions and
diseases (DI)
- Clinical trial status
(CL)
- Has mouse model (MO)
- Afftected biological process
(GO)
- Afftected cell types, tissues
(EX)
Drug Controlled Vocabulary Property Categories (and their respective codes)
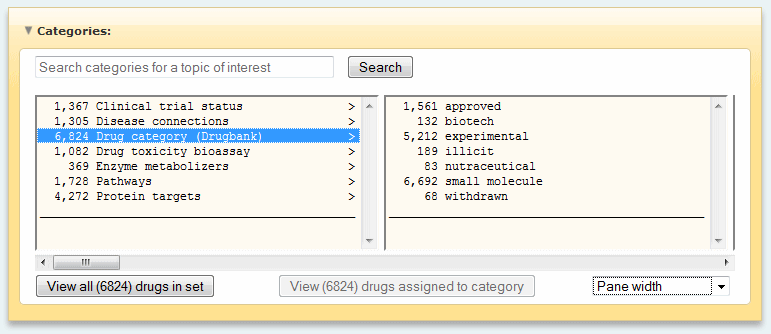
Drug properties
- Drug category (DrugBank)
(DC)
- Drug toxicity bioassay
(DT)
- Protein targets (PT)
- Clinical trial status
(CT)
- Disease connections
(DI)
- Pathways (PW)
- Enzyme metabolizers
(EM)
Please note: The terms in the driller may be sorted in ascending alphabetical order or in descending order by count (number of genes associated with a controlled vocabulary term or any child terms of the selected controlled vocabulary term). To change the sorting, select the desired option from the "Pane width" pull-down menu in the bottom right of the driller window.
Gene Controlled Vocabulary Category Descriptions
The following controlled vocabulary categories are used in the BKL.
Gene Ontology
The Gene Ontology classification system, developed by the Gene Ontology (GO) consortium, provides a comprehensive, logically hierarchical series of over 26,000 terms to describe genes by their Biological Process, Molecular Function, and Cellular Component.
Biological Process
Biological process terms describe the biological "objective" to which a protein contributes.
Molecular Function
Molecular function terms describe the biochemical activity of the protein.
Cellular Component
Cellular component terms describe the place in a cell where a protein is active.
Evidence Codes
GO properties are associated with an evidence code in the BKL. The evidence codes are:
BioKnowledge Transfer (K)
Indicates the information was predicted by BioKnowledge Transfer, a proprietary process that allows prediction of title lines and GO terms based on similarity to characterized proteins within the BKL (BLAST analysis) and on family membership and domain structure (Pfam analysis).
Experimental (E)
Indicates the information was demonstrated experimentally in published, peer-reviewed literature.
Predicted (P)
Indicates the information was predicted in published, peer-reviewed literature based on evidence other than sequence similarity, for example, association in a complex.
Sequence similarity (S)
Indicates the information was predicted in published, peer-reviewed literature based on sequence similarity to another protein.
Expression
Expression terms describe the spatial patterns of protein or mRNA expression during the development and maintenance of an organism. Expression information is available for human, mouse, rat, worm, and plant genes and is divided into four main categories:
- Expression Location
- Detection Technique
- Sample Information
- Expression Level
For human, mouse, and rat genes, over 300 Expression Locations terms describe the cell types, organs, tissues, fluids, and tumors where mRNA or proteins are expressed. Detection Technique, Sample Information, and Expression Level terms reflect the experimental method employed, the condition of the subject at the time the experiment was performed, and the degree to which the protein or mRNA was expressed relative to other samples.
For worm genes, Expression information is also available for genes represented in WormPD and is divided into two main categories: Expression Location and Developmental Stage. Over 500 Expression Location terms describe the cell types, organs, and tissues where mRNA or proteins are expressed. Developmental Stage terms reflect the temporal expression patterns for mRNA or proteins.
Plant Ontology
For plant genes, the BKL utilizes the Plant Ontology™ (PO) classification system, developed by the Plant Ontology Consortium (POC), which provides a comprehensive, logically hierarchical series of over 1,000 plant structure and developmental stage terms to describe genes and proteins.
Regulators of Fungal Genes
The controlled vocabulary for non-gene- or non-protein-based regulators of yeast gene expression. Regulator information is divided into two main categories:
- Regulators
- Effect of regulation
Regulators include environmental factors, chemical factors, cellular processes, and more. The Effect of regulation specifies whether the regulator induces, represses, or has no effect on the expression of the specified yeast gene.
Please note: Information regarding gene- or protein-based regulators is contained within the "Interaction with another protein" hierarchy.
Regulatory Factors and Targets
Search for genes included in TRANSFAC module, including transcriptional regulators and their targets.
Disease
The controlled vocabulary for Disease includes diseases (MeSH terms), human disease detail, and mouse models.
- Diseases are derived from MeSH terms, provided by the National
Library of Medicine.
- Disease terms may apply to human or mouse genes. When applied
to mouse genes, the mutant mouse is considered to function as a
model for the disease.
- Selecting the term "mouse models" after focusing on a disease
of interest limits the set of genes to mouse genes.
- Terms within the human disease detail hierarchy allow you to apply greater specificity to each of the disease terms including: degree of causality, presence and type of molecular alteration, form or stage of the disease, degree of association with tumors, etc.
Drug (DG)
The controlled vocabulary for Drugs is derived from DrugBank, which is the proximate source for drug entities contained in the BKL. The hierarchy is also divided into Pharmaceutical Type, which is subdivided into drug type, including:
- approved
- biotech
- experimental
- illicit
- investigational
- nutraceutical
- small molecule
- withdrawn
Phenotype
The controlled vocabulary for phenotype covers phenotypic changes for mutant mouse, yeast, worm, and plant genes in the Proteome module.
- Gross behavioral, physiological, and cellular phenotypes are
described for mouse knockout mutants.
- Gross morphological, behavioral, and growth phenotypes are described for null, loss-of-function, and gain-of-function S. cerevisiae, S. pombe, C. elegans, and pathogenic fungi mutants.
The phenotypes hierarchy is organized where the mouse phenotypes are considered separately from phenotypes of all other organisms. The following describes how the hierarchies are organized:
Mouse phenotype
Subdivided into anatomical phenotype, physiological phenotype, effect on viability, effect on fertility, experimental details, and affected anatomy.
The anatomical phenotype terms specify generally the effect of the phenotype, whether on the development, the function, or the composition of fluids. The actual location of the effect is specified in the affected anatomy terms. For example, a phenotype in liver development would have "change in development/morphology" as the anatomical phenotype and "liver" as the affected anatomy. Physiological phenotypes are divided into two general groups: cellular mechanisms, which are phenotypes that can be observed at the cellular level, and physiological processes, which are observed in the organism. Viability and fertility terms are grouped in the main list for convenience. Experimental detail terms reflect conditions under which phenotypes were observed, such as the gender of the mouse and the method the mutation was constructed with.
Other species (worm, fungi)
Subdivided into terms that are shared by more than one species (for example, lethal or conditional lethal), terms that are specific to one species or a subset of species, and terms which describe the mutation type. For example, follow Phenotypes > Other species (worm, fungi) > C. elegans phenotypes to browse those phenotypes specific to C. elegans. If you are interested in C. elegans and S. cerevisiae genes that produce a lethal phenotype when mutated, follow the path Phenotypes > Other species (worm, fungi) > Lethal or conditional lethal to the species non-specific term.
Plant phenotype
Trait Ontology™ (TO) terms, developed by Gramene, which provides a comprehensive, logically hierarchical series to describe traits and phenotypes associated with a gene or protein are used for curation of plant genes. Coupled with BIOBASE proprietary Mutation Type and Mutation Effect terms, TO terms form the basis of plant phenotype curation. In addition, Plant Ontology™ (PO) terms are used to capture structures and developmental stages affected by a mutation of a gene or protein.
Pathways and interactions
Pathway
Identifies genes which contribute to various canonical signaling and metabolic pathways
Interaction with other proteins
You may identify the set of genes whose protein products have a particular interaction effect with any interacting protein by selecting "Interaction with another protein" in the "Pathways and interactions" hierarchy of the Driller, then progressively drilling down to the desired manner of interaction (for example, binds directly, is directly regulated by, directly alters phosphorylation of).
Protein properties
Calculated properties: protein length, weight, pI, and predicted transmembrane segments
Protein length, weight, pI, and predicted transmembrane segments are calculated for all proteins. You may search for genes based on any of these properties using the predetermined ranges provided in the Driller, or by creating your own customized range. To use a predetermined range, highlight the range of interest (for example, protein length of 0-99 amino acids). To create a customized range, highlight one of the predetermined ranges, then enter your lower and upper limits into the two text boxes that appear in the Query frame.
Characterization
Describes the extent of experimental characterization of the protein. Proteins in the BKL are divided into three classes:
Characterized Proteins
Contain an experimentally determined Gene Ontology term (molecular function, biological process, or cellular component).
Uncharacterized Proteins
Have the title line, "Protein of unknown function" and have no GO properties other than "molecular_function_unknown", "biological_process_unknown", or "cellular_component_unknown".
Predicted Proteins
Include proteins that have undergone BioKnowledge® Transfer, which can result in the assignment of GO properties based on similarity to characterized proteins, and proteins that have properties assigned based upon predictions in the literature.
Family classification
A select list of gene families to which a protein may be assigned, based on the assignment of literature curated properties, as well as Pfam and BLAST analysis.
Pfam domains
Protein sequences in the BKL are compared to all Pfam domain/family seed sequences using HMMER, a multiple sequence alignment program using Hidden Markov Models. Family and domains that achieve the "trusted cut-off" score and are above the "noise cut-off" defined by Pfam for each Pfam seed sequence are displayed for each protein, referenced with the Pfam version used in the comparison. HMMER analysis, using the most recent version of Pfam, is performed approximately every two months, or as new versions of Pfam are released.
Post-translational modifications
The controlled vocabulary for post-translational modifications of proteins.
Species and Genetic Location
The species hierarchy includes information about the chromosomal location of the genes that encode the proteins in the BKL. To access the chromosomal location information, select "Species" in the far left panel of the Driller, then progressively drill down to the species of interest (Human, for example), then continue drilling down to the level of resolution desired for the chromosome of interest. Mouse is a special case where both cytogenetic and genetic position information is provided when available.
Orthologs Related to Current Set
Once you have focused on a set of mammalian genes, you may identify any mammalian orthologs related to the set of focused genes by selecting "Orthologs related to current set" in the far left panel of the Driller, then progressively drilling down to the group of species or species of interest (for example, human orthologs, mouse orthologs, or rat orthologs). Once you have selected the species of interest, the Query Frame will display the following options:
- Translate the current list of genes into the selected
species.
- Expand the list of current genes to include the corresponding
orthologs from the selected species.
- Limit the set of current genes to only those that have orthologs in the selected species.
Disease Controlled Vocabulary Category Descriptions
The following controlled vocabulary Property categories are used for diseases in the BKL.
Gene associations
Provides an alphabetized listing of proteins which are associated with one or more diseases in the BKL.
Co-occurring conditions and diseases
Provides a list of conditions or diseases (MeSH terms) that have been shown to be associated with other conditions or diseases.
Clinical trial status
Provides the status of clinical trials associated with the disease, as taken from ClinicalTrials.gov. To appear in the BKL, the "Condition" field for a given clinical trial must have been mapped to an existing BKL disease entry (MeSH term).
Has mouse model
Indicates that a knock-out mouse model system has been used in the study of the disease.
Affected biological process
Provides a list of biological processes (taken from the Biological Process hierarchy provided by the Gene Ontology (GO) consortium) that have been shown to be affected by a condition or disease.
Affected cell types, tissues
Provides a list of cell types and tissues (taken from BIOBASE's internally developed expression ontology) that have been shown to be affected by a condition or disease.
Drug Controlled Vocabulary Category Descriptions
The following controlled vocabulary Property categories are used for drugs in the BKL.
Drug category (DrugBank)
Indicates the category that a drug has been assigned to by DrugBank.
Drug toxicity bioassay
Indicates toxicity bioassays that a drug has been investigated for as reported by the FDA and published in the following reference:
Matthews, E.J., Kruhlak, N.L., Cimino, M.C., Benz, R.D., and Contrera, J.F. An Analysis of Genetic Toxicity, Reproductive and Developmental Toxicity, and Carcinogenicity Data: I. Identification of Carcinogens Using Surrogate Endpoints. Regulatory and Toxicology and Pharmacology, 2006.
Protein targets
Provides an alphabetized listing of proteins which are listed as the target of one or more drugs at DrugBank.
Clinical trial status
Provides the status of clinical trials associated with the drug, as taken from ClinicalTrials.gov. To appear in the BKL, the "Indication" field for a given clinical trial must be of type "Drug" and must have been mapped to an existing BKL drug entry (taken from DrugBank).
Diseases connections
Provides a list of conditions or diseases (MeSH terms) that have been for which the drug is under investigation as a treatment, as taken from ClinicalTrials.gov. To appear in the BKL, the "Indication" field for a given clinical trial must be of type "Drug" and must have been mapped to an existing BKL drug entry (taken from DrugBank).
Pathways
Provides a list of pathways which a drug may be associated with, via its interaction with a protein target or metabolizing enzyme.
Enzyme metabolizers
Provides an alphabetized listing of proteins which are listed as the metabolizing enzyme of one or more drugs at DrugBank.