Creating Custom Models with the Composite Model Editor
Use the Composite Model Editor, shown below, to create models. The term "model" describes a specific pair of weight matrices from TRANSFAC with matrix similarity cut-off values for each matrix.
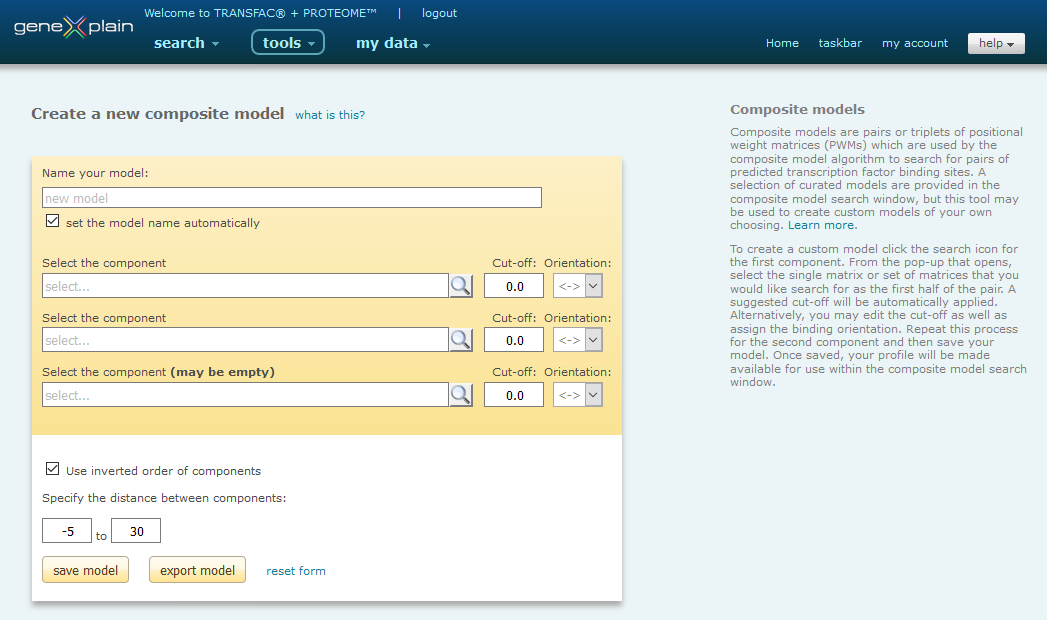
Composite Model Editor
Create a Model
Creating new model using Composite Model Editor involves the following steps:
- Select the first and second components of the
pair
- Set optional parameters (if
desired)
- Save the model
1. Select the First and Second Components of the Pair
To create a new model, first select the matrices to represent each component.
To select a matrix, click the search button in the "Select the component" box. A pop-up window will open which contains the complete contents of the TRANSFAC matrix library plus any user uploaded matrices.
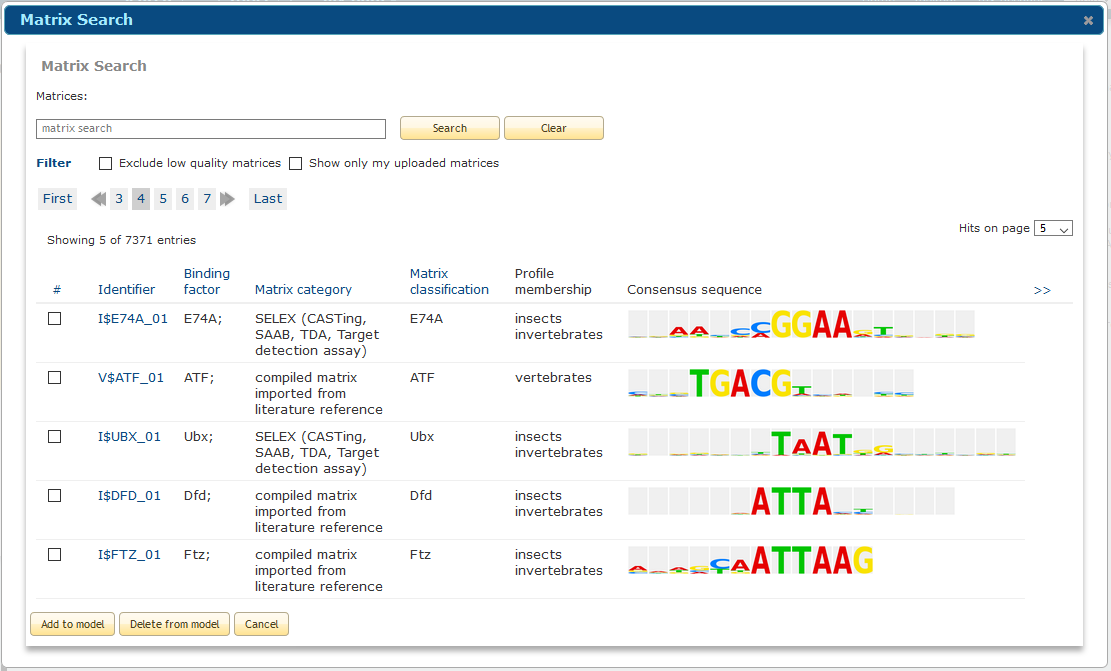
Matrix selection view
Search for matrices to select by typing accession number, matrix identifier or factor/matrix name (or a part thereof) in the search box above the matrix list and click Search. Use the "Exclude low quality matrices" option to hide all matrices of low quality. Use the "Show only my uploaded matrices" to restrict the view to only those matrices which you have uploaded.
When you have selected one or more matrices to represent the component, click the "Add to model" button. The matrix or matrices that you have selected will be automatically added to the model. The minFN cut-off is selected by default, and is recommended for the composite model analysis but you have the option to specify the cut-off directly. The default orientation is set to not consider the direction, but you may set any direction as desired.
Repeat this process for the second component.
2. Set optional parameters (if desired)
Two optional parameters may be set:
Use inverted order of components
By default, when the "Use inverted order of components" option is selected, the order in which the components are identified within the submitted sequence will not be considered when determining a match. For example, if Matrix A is selected as component 1 and Matrix B is selected as component 2, the model will be returned in the results regardless of whether the order is A -> B or B -> A, as long as the cut-off criteria are met. When the "Use inverted order of components" selection is turned off, the order in which the components are identified will determine whether a match is made. For example, if Matrix A is selected as component 1 and Matrix B is selected as component 2, and "Use inverted order of components" is turned off, only A -> B models will be returned in the results when the cut-off criteria is met. B -> A models will be ignored.
Specify the distance between components
This parameter specifies the maximum distance, in nucleotides, that may exist between the two components in order for a pair of binding sites to be identified. A negative starting distance specifies the number of nucleotides by which the two matrices may overlap. A default distance of -5 to 50 is set, which is appropriate for most analyses.
3. Save the model
To save the model, keep the suggested name or enter your own name by de-selecting the "set the model name automatically" option. The newly-created model will be displayed and can immediately be used in Composite Model search. The new model will be listed in the Composite Model main interface in the model selection list as well as in the Gene regulation analysis -> Data -> Composite models folder of the my data Data Management system.
Manage Existing Models
View a Model
Created models can be viewed by selecting the desired model from the Gene regulation analysis -> Data -> Composite models folder of the my data Data Management system.
Delete a Stored Model
Click the word "delete" next to the model that you wish to delete in the Gene regulation analysis -> Data -> Composite models folder of the my data Data Management system.