miRNA Analysis (TRANSFAC)
The miRNA analysis is available from the tools menu.
Putative gene targets are assigned to miRNAs based on pre-calculated total context scores from TargetScan (http://www.targetscan.org/). The tool compares for each miRNA the number of potential targets in the analysis data set against the number of potential targets in the background set selected by the user. The enrichment of the gene targets in the analysis is calculated by dividing the relative number of hits in the analysis set by the relative number of hits in the background set. P-values for significantly enriched targets are calculated based on a hypergeometric distribution. As cut-off a p-value of 0.01 is applied.
The miRNA analysis is also available as part of the Step-by-step data analysis of gene-based microarray and RNA-seq data sets. The miRNA analysis result can be accessed via the link on the top of the respective FMatch Report. If the FMatch Report is saved, the miRNA result is saved as well.
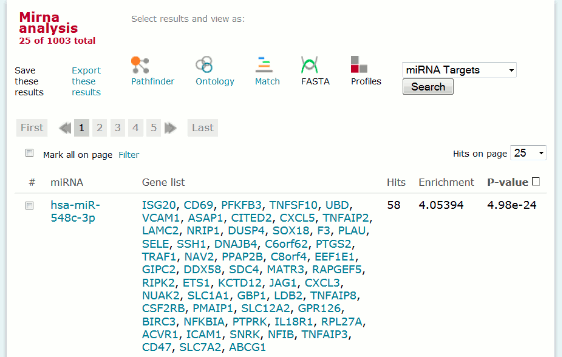
The miRNA result is displayed in form of a database search result with the following columns:
- miRNA for which the fraction of targets among the compared gene sets differs significantly (p-value < 0.01)
- List of target genes (hits) among the analyzed genes
- Number of hits (a) among the analyzed genes (A)
- Enrichment = (a/A)/(b/B), where A = number of genes in the analysis set; a = number of hits/target in the analysis set; B = number of genes in the background set; b = number of hits/targets in the background set
- Hypergeometric p-value
Complete or filtered lists of the miRNAs in the result can be exported or subjected to further analysis using the functions on top of the result table.
Contact us at support@genexplain.com