Reaction Hierarchy
The Reaction Hierarchy shown in Figure 1 was introduced in release 5.1 to connect the patchwork of molecular evidence reactions (different species and modified forms in the various experiments) to a mechanistic level that models pathways. The pathway level consists of direct pathway step reactions, indirect pathway step reactions (in cases where the direct mechanism for a part of the pathway is not known) and auxiliary decomposition reactions. In addition, there are metabolic pathway step reactions. In combination with reaction chains, this level is used to model canonical pathways and networks.
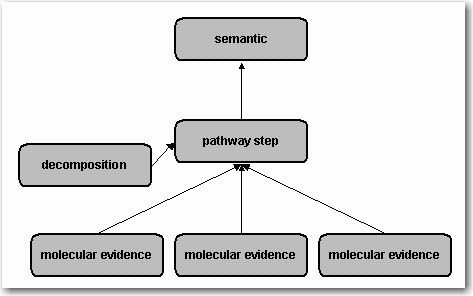
Figure 1. Reaction Hierarchy
In contrast, the semantic level gives a broad overview of the signal transduction pathways without biochemical details. Detailed desription of the mechanistic and semantic levels are provided below.
Mechanistic Reactions: Molecular Evidence, Pathway Step, Decomposition
The classical chemical reaction notation shown in Figure 2 can model aggregation into a complex, dissociation of a complex, chemical modification and catalysis in the detail necessary to capture data from the primary literature.

Figure 2. Classical Reaction Notation
An example of mechanistic reaction data that can be captured using classical chemical reaction notation is shown in Figure 3.
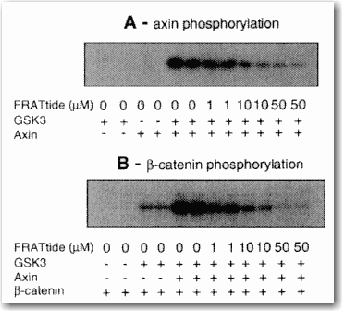
Figure 3. Example of Mechanistic Reaction from the Primary Literature The data show that B-catenin and axin are phosphorylated by GSK3, and the presence of axin increases the phosphorylation of B-catenin. FRATtide inhibits the phosphorylation by competition [C. M. Thomas et al. FEBS Letters, 458:247–251, 1999]. Another paper [H. Aberle et al. EMBO J., 16:3797–3804, 1997] then shows that the phosphorylated form of B-catenin is ubiquitinated and degraded by the proteasome.
This representation is referred to as mechanistic in BKL. The mechanistic format illustrates how the molecules interact and not how the signal is transduced. This format is used for the pathway step, decomposition and molecular evidence reaction types in BKL.
Molecules can act as enzymes, substrates, modulators, or products, and any reaction has no more than one enzyme, one or more substrates, any number of modulators, either inhibitors or activators, and one or more products. There is semantic information in assigning some of the inputs to the enzyme or modulator collections, which tells us something about the local meaning of the molecule for the reaction.
This view requires detailed knowledge of the mechanisms of transduction. It makes no assumptions about the molecule states.
Semantic Reactions
A representation of format of reactions commonly encountered in scientific reviews is shown in Figure 4. This format is referred to as semantic in BKL. The semantic format shows how the signal flows through the network but does not provide detail about whether the molecules activate or inhibit the target molecule.
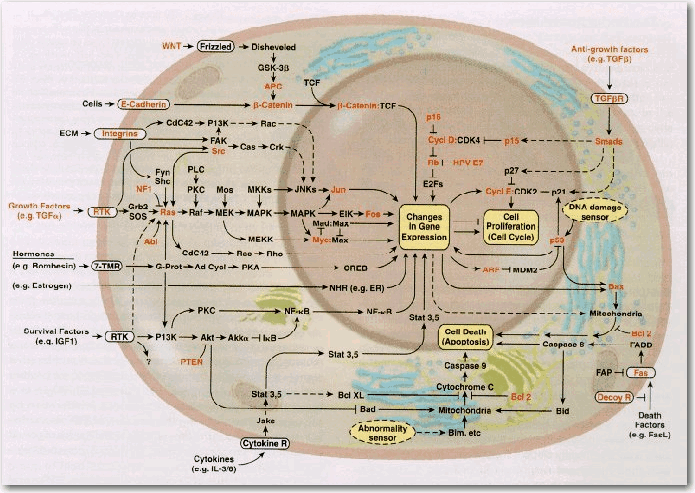
Figure 4. Example of Semantic Reactions Depiction of a cancer cell signaling network [taken from: D. Hanahan and R. A. Weinberg, The hallmarks of cancer; Cell, 100(1):57–70 Review, 2000]. Lines with arrows indicate activating reactions. Lines with bars indicate inhibiting reactions.
The semantic view assumes that each molecule exists in an "active" state and an "inactive" state. It is commonly understood that incoming activating reactions always refer to the inactive state, while incoming inactivating reactions and outgoing reactions refer to the active state. Because the states are implied for semantic reactions, we link them to protein molecules in BKL.
Saying that a molecule has an "active" or "inactive" state is a semantic statement. Both states undergo reactions, and the decision as to which is which can consequently only be determined in the larger context of the whole network.
Please note: In a translocation, the same molecule enters and leaves a reaction, with a changed spatial context. This process takes some time, which is important for the dynamic behavior of the network. Translocations cannot be represented in the basic mechanistic model that assumes all reaction partners are present in the same reaction space. Since a molecule is just associated with a list of locations, we cannot differentiate between, for example, the cytosolic and the nuclear form of the molecule. A reaction that moves a molecule from one form to the other is then a loopback from the molecule to itself. Therefore, we associate the two locations with the connection entries between the molecule and the reaction. The term translocation is assigned in the effect (EF) field.
Click here for information about the format for Reactions in BKL.