Genes and Molecules Described in the BKL
The important characteristics of components of a group are what define those components as members of the group and allow researchers to identify relationships between and among the members. For example, molecules with certain shared characteristics are grouped into families or superfamilies. Similarly, reactions with shared characteristics can be assigned into general reaction classes, as has been done in the EC nomenclature. Lastly, combinations of molecules and reactions can be grouped, and the grouping may correspond to a particular network or pathway.
Genes and Molecules in the BKL are grouped into the following categories:
Genes
Genes and Orthogenes
Gene describes a single gene from a particular taxon. The prefix ortho is added (orthogene) to describe a group of orthologous genes from multiple species or higher taxons. Genes and orthogenes are described on Locus Reports.
Proteins
Family and Orthofamily Molecules
Grouping molecules into families and orthofamilies allows us to write algorithms that exploit these relationships and infer properties for the individual family members. Molecules from a single species or higher taxon that comprise a family or superfamily are grouped into a family. Orthofamily describes a group of homologous families or superfamilies from multiple species or higher taxons. Families and orthofamilies are described on Family Reports.
Isogroup and Orthogroup Molecules
Several isoforms may exist for a particular gene product. Occasionally, a signaling activity is attributed to a single gene product, and researchers discover later that several molecules are produced by that gene. The BKL has molecule categories that refer to the products of a particular gene, where the particular isoform is not known or specified in the scientific literature. Isogroup describes a group of species- or higher taxon-specific products of a single gene. Orthogroup describes a group of products from orthologous genes from multiple species or higher taxons. Isogroup and orthogroup molecules are described on Locus Reports.
Protein and Orthobasic Molecules
Often the isoform, a particular splice variant for example, is specified in the scientific literature. Protein describes a specific isoform from a particular species or higher taxon. Orthobasic describes a group of specific isoforms produced by orthologous genes from multiple species or higher taxons. Proteins and orthobasic molecules are described on Locus Reports.
Complexes
Complex and Orthocomplex Molecules
Non-covalently bound molecules from a particular species or higher taxon are referred to as a complex. Homologous complexes from multiple species or higher taxons are referred to as an orthocomplex. Complexes and orthocomplexes are described on Complex Reports.
The figure below illustrates the relationships between and among various genes, proteins, and complexes in the BKL.
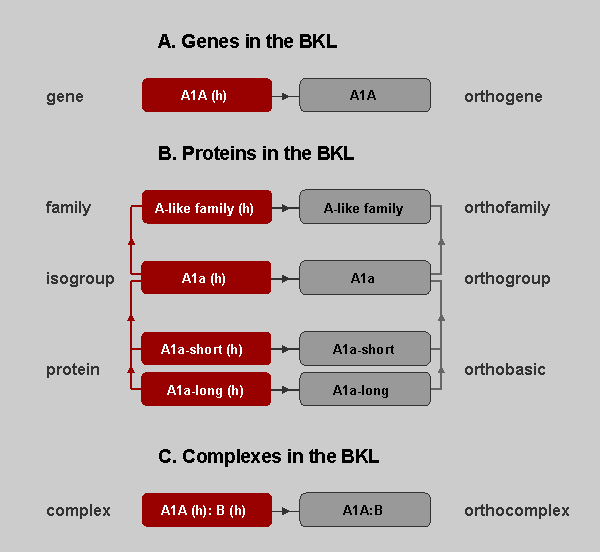
Genes, Proteins, and Complexes in the BKL. Types that describe groups of orthologous genes and molecules appear in gray. Types that specify particular taxons appear in red, with taxon designated in parentheses. Here, (h) indicates human. Arrows indicate that the gene or molecule is a component of a particular group. A. Genes in the BKL. Genes and orthogenes are described on Locus Reports. B. Proteins in the BKL. Family and orthofamily molecules are described on Family Reports, while isogroup, orthogroup, orthobasic, and protein molecules are described on Locus Reports. C. Complexes in the BKL. Complexes are indicated with colon separating the individual components (A1A:B). Complex and orthocomplex molecules are described on Complex Reports.
Small Molecules
Non-proteinaceous molecules that play regulatory roles in signaling pathways are also described in the BKL, on Small Molecule Reports.
Preserving Scientific Context with Molecule
Types in TRANSPATH
Examining the sequence of a gene or protein is easier than
investigating its function. Typically, certain characteristics
are experimentally determined for a few members of a protein
family, and homologs are added to the functional group by sequence
or structural similarity. Various databases classify proteins
and map sequence motifs to functional annotation using this
premise. They cluster proteins by multiple sequence alignments and
use common structural motifs, "profile" patterns, or Hidden Markov
Models derived from these alignments to classify new proteins.
Sometimes these methods can correctly predict function, but
sometimes not. Thus, it is common practice to group molecules into
families on the basis of sequence similarities, even if they do not
share common characteristics.
For TRANSPATH, it is advantageous to group molecules that show
common signaling characteristics. Since we are primarily interested
in function, we group molecules by function. In addition, we want
to draw from information derived from expert knowledge and remain
consistent with groupings that already exist. To solve this
dilemma, we group the molecules as it is done traditionally, but
link signaling only to those molecules for which it has been shown
experimentally.
Given the relationships described in the figure above, if a
reaction has been demonstrated for A1a(h), we link that reaction to
the A1a(h) molecule only. We link statements made on a general
level, such as those derived from reviews, for example, to nodes on
a higher level in the molecule hierarchy. For example, we can link
the general activation of A-like proteins to the orthofamily entry
"A-like family", and the context of the original statement in the
scientific literature is preserved.
Additional details about the TRANSPATH data model that has been used to describe molecule types in the BKL can be found at:
Choi C, Crass T, Kel A, Kel-Margoulis O, Krull M, Pistor S, Potapov A, Voss N, Wingender E. (2004) Consistent re-modeling of signaling pathways and its implementation in the TRANSPATH database, Genome Inf. Ser. 15: 244-254. [Abstract].